AI’s Power in Diagnostics and the Hunt for New Antibiotics
Artificial intelligence (AI) is rapidly emerging as a transformative force in the battle against infectious diseases and antimicrobial resistance (AMR), a critical global health threat. Traditional approaches to diagnosis and antibiotic discovery are often slow and costly, struggling to keep pace with the evolution of drug-resistant microbes. This paper explores the profound difference AI is making in disease control and the quest for new antibiotics. It will delve into the specific AI methodologies employed, the types of data leveraged and their rationale, the key findings, inherent limitations, and the overarching long-term value of AI in this crucial domain.
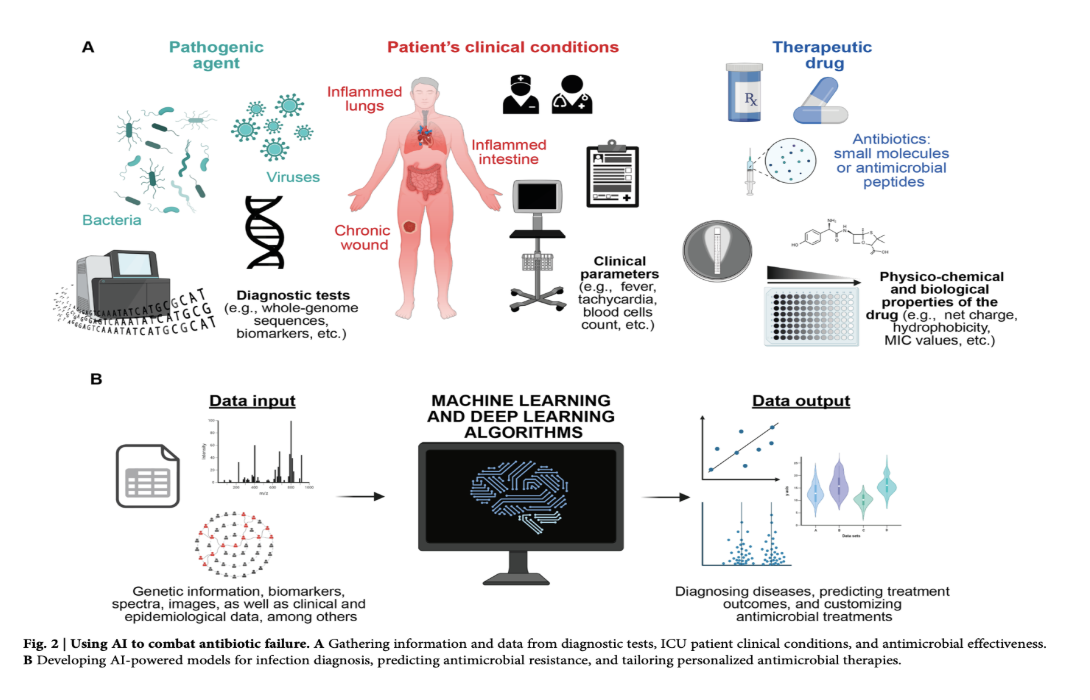
AI is revolutionizing disease control, particularly in diagnostics and treatment strategies. In diagnostics, conventional tests often delay crucial interventions, especially in critical conditions like sepsis. AI-driven methods, leveraging machine learning (ML) and deep learning (DL), are enhancing pathogen detection and AMR prediction. For instance, convolutional neural networks (CNNs) have been adapted to accurately identify bacteria from Gram stain images with approximately 95% accuracy. Similarly, CNNs are being used to differentiate susceptible from resistant bacterial phenotypes at the single-cell level. Recurrent neural networks (RNNs) and long short-term memory (LSTM) networks are employed to analyze time-series data from intensive care units (ICUs) for early detection of bloodstream infections and sepsis. The data used in these diagnostic applications ranges from microscopy images and mass spectra from MALDI-TOF MS to bacterial whole-genome sequences and clinical parameters from EHRs. The purpose of analyzing this diverse data is to identify complex patterns indicative of infection and resistance, often surpassing the capabilities of traditional statistical methods.
AI is also advancing personalized treatment. ML models analyzing extensive EHR data, including demographics, medical history, and drug purchase records, can predict antibiotic resistance on an individual basis, leading to a significant reduction in mismatched prescriptions. For example, an ML model analyzing UTI patient records outperformed clinicians, slashing second-line antibiotic usage by 67%. Furthermore, AI-powered chatbots are being used to disseminate timely information about infectious diseases, symptoms, prevention, and treatment options, improving patient education and access to care. These chatbots utilize techniques like pattern matching, ML (decision trees, random forests), and natural language processing (NLP) to understand and respond to user queries.
In antibiotic discovery, AI offers the potential to dramatically accelerate the identification of novel antimicrobial compounds. Traditional drug discovery is a lengthy and expensive process. AI bypasses some of these limitations through in silico virtual screening of vast chemical libraries and the prediction of efficacy and safety. Quantitative structure-activity relationship (QSAR) models, utilizing various ML algorithms like support vector machines (SVMs) and neural networks, predict the biological activity of small molecules and antimicrobial peptides (AMPs) based on their structural and property descriptors. Deep learning models, including message-passing neural networks (MPNNs) and graph neural networks (GNNs), are trained on large datasets of molecules to predict antibacterial activity. Generative deep learning models are employed to design novel molecules with desired characteristics, even exploring chemical spaces beyond known compounds. These models are trained on vast databases of drug-like molecules or bioactive peptides. Findings include the AI-guided discovery of new antibiotics like halicin and abaucin, and the computational identification of thousands of potential AMPs from the human proteome and even extinct organisms through "molecular de-extinction". AI is also being used to predict biomolecular properties and structures, aiding in understanding the mechanisms of action of potential drugs, with tools like AlphaFold predicting protein structures with high accuracy.
Despite these advancements, there are limitations. In diagnostics, AI models require high accuracy to be clinically reliable, and false positives or negatives can have serious consequences. The "black-box" nature of some DL algorithms raises concerns about interpretability and trust. Data availability and quality remain significant challenges across all applications, with biases in datasets potentially leading to unfair or inaccurate outcomes for certain patient groups. In antibiotic discovery, while AI can generate numerous potential candidates, many may be difficult to synthesize or may not possess the necessary secondary properties for clinical viability, such as appropriate pharmacokinetics and low toxicity. Ethical concerns surrounding data privacy, informed consent, and algorithmic bias must also be carefully addressed.
In conclusion, the long-term value of AI in infectious disease control and antibiotic discovery is immense. AI's ability to analyze vast, complex datasets, accelerate diagnosis, personalize treatment, and expedite the identification of novel antimicrobials offers a powerful arsenal in the fight against AMR. As AI technology continues to advance, and as data availability and quality improve, we can anticipate more precise diagnostic tools, optimized treatment strategies, and a strengthened preclinical antibiotic pipeline. The integration of AI into clinical microbiology laboratories and the development of interpretable models will further enhance its utility. However, realizing the full potential of AI requires addressing current limitations, upholding ethical principles, and fostering collaboration among healthcare professionals, developers, and policymakers. Ultimately, AI is not a silver bullet but a crucial tool that, when responsibly developed and implemented, holds the promise of significantly mitigating the threat of infectious diseases and AMR.
If you or your team is ideating on the best ways to apply AI to your life sciences work, please reach out to KAMI Think Tank (info@kamithinktank.com).
References
https://www.nature.com/articles/s44259-024-00068-x?fromPaywallRec=false